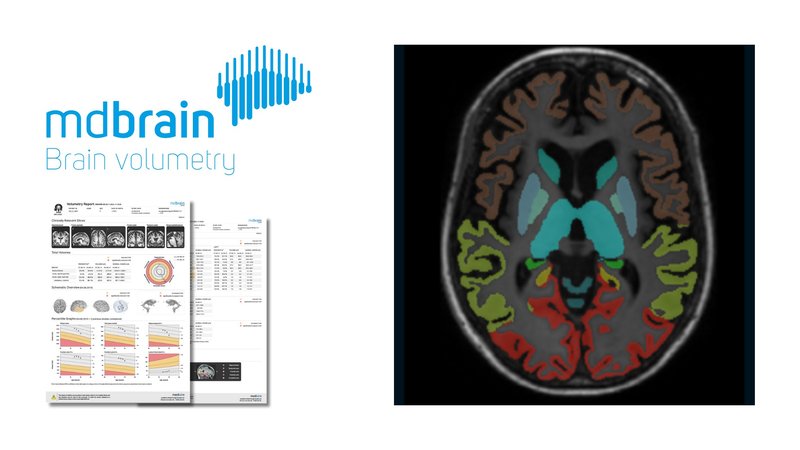
In a retrospective study published in Scientific Reports, researchers used the deep learning-based software, mdbrain by mediaire, for automated volumetry to assess brain atrophy in patients with Huntington’s disease. This evaluation aimed to validate the software's efficacy against manual measurement techniques commonly used in clinical settings. The study analyzed twenty-two patients, including a control group, matched by age and sex. The findings revealed that mdbrain’s automated volumetry could significantly differentiate between the patient and control groups, showing only a -2.3% mean relative discrepancy in caudate nucleus volume measurements. The certified software demonstrated its capacity to objectively quantify atrophy in several brain structures, offering considerable improvements over standard semiquantitative methods. This external validation study highlights how automated tools like mdbrain can enhance diagnostic accuracy and efficiency in clinical practice, particularly for neurodegenerative diseases such as Huntington’s.
Read full study
External evaluation of a deep learning-based approach for automated brain volumetry in patients with huntington’s disease
Scientific Reports, 2024
Abstract
A crucial step in the clinical adaptation of an AI-based tool is an external, independent validation. The aim of this study was to investigate brain atrophy in patients with confirmed, progressed Huntington's disease using a certified software for automated volumetry and to compare the results with the manual measurement methods used in clinical practice as well as volume calculations of the caudate nuclei based on manual segmentations. Twenty-two patients were included retrospectively, consisting of eleven patients with Huntington's disease and caudate nucleus atrophy and an age- and sex-matched control group. To quantify caudate head atrophy, the frontal horn width to intercaudate distance ratio and the intercaudate distance to inner table width ratio were obtained. The software mdbrain was used for automated volumetry. Manually measured ratios and automatically measured volumes of the groups were compared using two-sample t-tests. Pearson correlation analyses were performed. The relative difference between automatically and manually determined volumes of the caudate nuclei was calculated. Both ratios were significantly different between the groups. The automatically and manually determined volumes of the caudate nuclei showed a high level of agreement with a mean relative discrepancy of - 2.3 ± 5.5%. The Huntington's disease group showed significantly lower volumes in a variety of supratentorial brain structures. The highest degree of atrophy was shown for the caudate nucleus, putamen, and pallidum (all p < .0001). The caudate nucleus volume and the ratios were found to be strongly correlated in both groups. In conclusion, in patients with progressed Huntington's disease, it was shown that the automatically determined caudate nucleus volume correlates strongly with measured ratios commonly used in clinical practice. Both methods allowed clear differentiation between groups in this collective. The software additionally allows radiologists to more objectively assess the involvement of a variety of brain structures that are less accessible to standard semiquantitative methods.
